PRT Classification Objects
One of the most powerful features of the Pattern Recognition Toolbox is classification objects, implemented as prtClass objects. Classification objects allow you to develop algorithms which will label data into discrete clases. prtClass objects are all supervised, meaning they require labeled training data during training.
Contents
Classification object methods and properties.
All prtClass objects inherit the TRAIN, RUN, CROSSVALIDATE and KFOLDS functions from the prtAction object, for more information on these methods, refer to section on the prtEngine.
In addition to the inherited methods, prtClass objects also have a few important properties. The isNativeMary field indicates whether or not the particular classifier natively handles binary and/or M-ary classification. Binary classifiers can only label data as being in class 0 or 1, whereas native M-ary classifiers can label data into an arbitrary number of classes.
Using classifiers
You use classifiers in the same manner as any prtAction object. The following example shows how to create a generalized likelihood ratio classifier, and perform kfolds validation on it.
ds = prtDataGenUnimodal; % Load a dataset to use classifier = prtClassGlrt; % Create a generalized likelihood ratio test % classifier result = classifier.kfolds(ds,2);% Perform a simple 2-fold cross-validation result.getX(1:5) result.getY(1:5)
ans =
-7.0771
1.6891
-15.2419
-7.3593
-10.2619
ans =
0
0
0
0
0
Note that the data stored in the observations of result correspond to the likelihood values. Also note that since ds was a labeled dataset, the original labels are copied over into the targets property of the results dataset.
Internal Deciders
Another important property of prtClass objects is the internalDecider. Ordinarily, a prtClass object outputs raw statistics based on the classification algorithm. However, you might just want the classification object to make class decisions based on these outputs. This can be done by setting the internalDecider property to be a prtDecisionBinaryMinPe object:
classifier.internalDecider = prtDecisionBinaryMinPe; result = classifier.kfolds(ds,2); %Perform a simple 2-fold cross-validation result.getX(1:5) result.getY(1:5) % Note that now the data stored in the observations of result are class % labels. They are likely all of class 0 in this example. By setting the % internalDecider to prtDecisionBinaryPe, an threshold was found during % training that would result in the minimum probability of error.
ans =
0
1
0
0
0
ans =
0
0
0
0
0
Plotting
Finally, prtClass objects all have an additional plot function, which can help you visulize the classifiers decision regions. To plot the classification object, it first needs to be trained.
classifier = classifier.train(ds); % For example purposes, % train with all the data classifier.plot(); % Alternatively, plot(classifier)
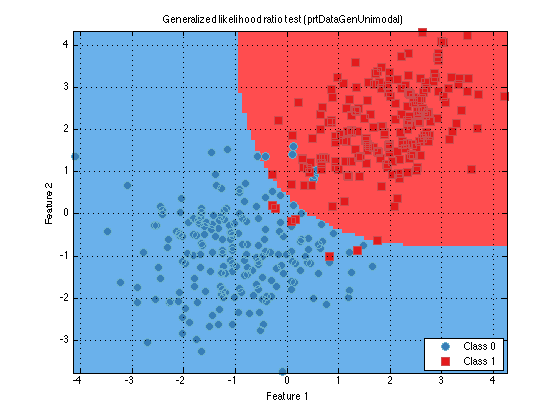
In the resulting plot, you will see all the data members used to train the data. If the internalDecider is set, as in the above example, you will see the decision region boundaries. If the internalDecider is not set, you will instead see an intensity plot, indicating how likely it is that a particular point would belong to class 0 or 1, as shown below.
classifier.internalDecider = []; % Clear the internalDecider classifier = classifier.train(ds); % Re-train classifier.plot()
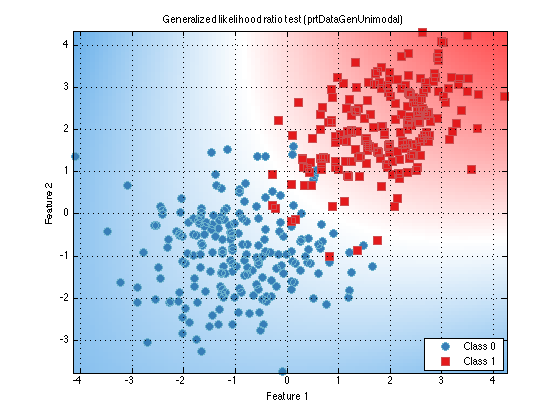
All classification objects in the Pattern Recognition Toolbox have the same API as discussed above. The only difference is the underlying algorithms used to train and run the classifier. For a list of all the different classification algorithms, and links to their individual help entries, A list of commonly used functions